It swept across the planet, less than 100 years ago, like the Black Plague: A few people living today still remember the coffin shortages, the overflowing hospital wards, the friends who were alive one week and gone the next. In 1918, the so-called Spanish flu blanketed a world at war with a second devastation, killing more than 20 million people in a few short months. What made the Spanish flu such a killer? A report in Friday’s issue of the journal Science suggests that the 1918 pandemic may have been sparked by an unusual recombination of pig and human flu genes.
There have been other flu outbreaks in the past century, including severe epidemics in 1957 and 1968, and a short-lived scare with the Hong Kong chicken flu in 1997. But none has rivaled the virulence of 1918 flu.
The detective story behind the 1918 outbreak would have been impossible to write just a few short years ago, because the story’s central focus — the virus itself — was unknown. With no living copies of the virus available to study, enterprising researchers located and recovered fragments of the flu genome in carefully preserved and archived lung tissues from World War I soldiers, and in one case, from an Inuit woman buried in the Alaskan permafrost.
The RNA sequences recovered from these time-capsule tissues confirmed that the 1918 flu was a variant of the H1N1 subtype of influenza, versions of which still circulate among humans today. Early analyses suggested that the flu was most closely related to mammal flu viruses, and that the ancestors of the virus must have infected mammals several years before 1918. Yet this first look at the 1918 flu yielded few clues about its unusual virulency.
A lethal combination
Flu viruses are basically strands of RNA, enveloped in a layer of fat and studded on the surface with spikes of hemagglutinin and neuraminidase proteins. Hemagglutinin helps the virus attach to host cells, while neuraminidase helps the viral particles eventually escape from the host cell and spread the infection. The body develops antibodies against these two proteins to protect itself from flu.
The genes behind these proteins are constantly mutating, however, gradually taking on different “looks” that existing antibodies find harder to recognize and combat. This never-ending cycle is the reason why new flu vaccines must be created every year, to match the new variants that have sprung up since the last flu season.
In some cases, new versions of these genes abruptly appear, and the virus’ hosts are suddenly faced with a threat against which they have no antibodies. The introduction of bird hemagglutinin genes into human flu strains before the 1957 and 1968 flu epidemics, for instance, show how these sudden shifts can produce new virulent strains.
New viral genes can also arise through a process called recombination, where pieces of a gene from two or more parent viruses combine to form one new gene. Research by Mark J. Gibbs, his father Adrian J. Gibbs, and John A. Armstrong of the Australian National University on plant viruses suggests that recombination can create especially virulent strains. Although recombination had never been seen in flu genes, the team decided to see if recombination posed a similar threat among those viruses.
“We talked about looking at flu sequences, but we didn’t know where to start with flu because there are so many sequences, more than 6,000, and recombination in flu was thought to be very rare,” said Mark Gibbs.
Then, one night, Adrian Gibbs came across an article about the newly recovered 1918 virus and decided to examine it for signs of recombination. In a few hours, he had found an unusual signal in the sequence data.
“He couldn’t sleep until he told me about it the next day,” said Mark Gibbs.
Pig and human
The data in question was the complete sequence for the 1918 flu’s hemagglutinin gene. After exhaustively comparing the sequences with hemagglutinin sequences from a variety of bird, pig, and human flu types, the researchers concluded that the 1918 hemagglutinin was a chimera, a true recombinant gene made up of sequence from a pig flu virus and a human flu virus.
Their analysis showed that the beginning and end parts of the gene sequence came from human flu, while the middle part of the gene sequence came from pig flu. These different regions of the gene encode different parts of hemagglutinin: The ends encode the stalk that attaches the protein to the viral coat, while the middle encodes hemagglutinin’s globular head. Mark Gibbs suggests that this relatively neat breakdown may have been a factor in the recombinant gene’s survival.
“This is the first known case where a viable gene has been produced from the parts of other influenza genes,” said Mark Gibbs.
In addition to its unusual virulence, the 1918 flu was notorious for killing an extraordinary number of young adults. Younger people were perhaps more vulnerable to the flu, the new research suggests, because their immune systems had never encountered the new recombination, and thus couldn’t recognize or fight it.
Did the recombination of the hemagglutinin gene actually launch the flu pandemic of 1918? The pig/human gene emerged about the same time as the pandemic, suggesting that, indeed, genetic reshuffling may have triggered this public-health disaster.
“The start of the pandemic coincided with a recombination event that might produce the phenotypic novelty required to trigger a pandemic,” the Science article concludes. “This coincidence suggests a causal link.”
Moreover, recombination “can result in increased virulence,” Gibbs says.
A second study in the same issue of Science seems to support this notion: In mice, a single amino acid substitution in the PB2 gene and hemagglutinin gene strongly influences infection rates of the 1997 Hong Kong chicken flu. This report, by Yoshihiro Kawaoka of the University of Wisconsin-Madison and the University of Tokyo, with Masato Hatta and others, is “consistent with the concept that influenza virus pathogenicity is multigenic.”
Could it happen again?
Though such evidence seems convincing, it’s still not possible to say with absolute certainty whether a change in the hemagglutinin gene gave the 1918 flu its extra kick, Graeme Laver of Australian National University and Elspeth Garman of the University of Oxford say in a related essay. The bigger question now, they write, is whether similarly virulent flu strains are likely to strike again — and whether modern medicine will be ready in the event of a new pandemic.
Mass vaccination, as attempted in 1976 when a swine flu outbreak occurred among army recruits at Fort Dix, N.J., can be problematic, Laver and Garman say. In that case, the preventive efforts were undermined by vaccine side effects and litigation tangles, as well as disappointing antibody responses.
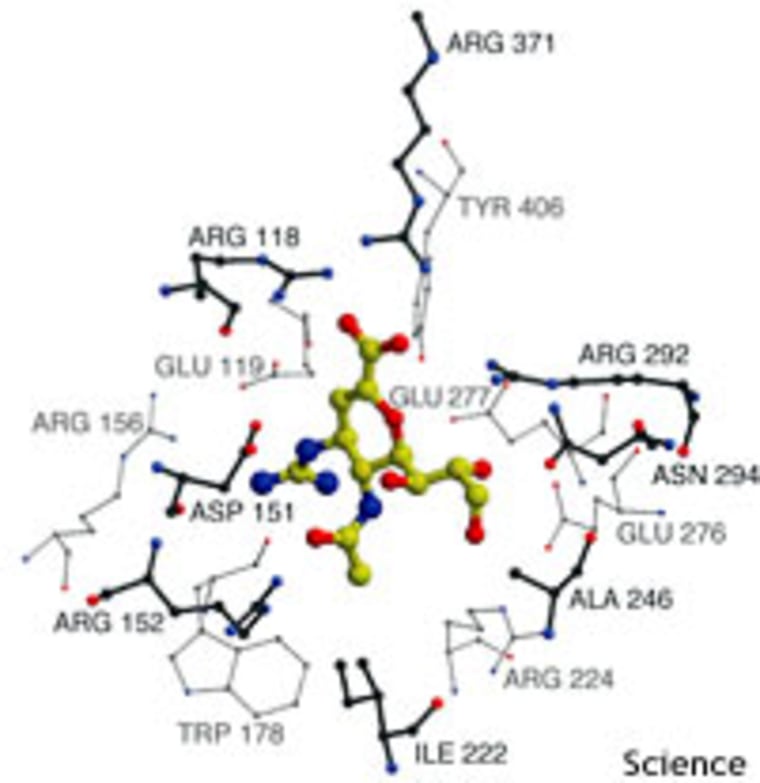
Current flu vaccines, containing hemagglutinin and neuraminidase from various strains of cultured flu virus, only target particular versions of the virus. Developing and safety-testing enough vaccine to fight a new virus would take time — perhaps six months — as well as high levels of research investment.
The most promising “first line of defense,” Laver and Garman propose, is antiviral drugs, particularly neuraminidase inhibitors. But, they point out, “for the drugs to be of any use, huge quantities would need to be immediately available, and means for their rapid distribution would need to be in place beforehand.”
Unfortunately, current supplies are “woefully inadequate,” Laver and Garman say. Robert G. Webster of St. Jude Children’s Research Hospital echoes this sentiment: “During the period between detection of a pandemic strain and the availability of a vaccine,” Webster writes, “antiviral drugs will be essential. It is gravely disquieting that no action has yet been taken to create strategic stockpiles of such drugs.”